Overvåking og studier gjennomført ved Havforskningsinstituttet og Veterinærinstituttet har aldri avdekket meldepliktig sykdom i norske flatøsters, Ostrea edulis. I 2006 ble det observert mikroskopiske celler som liknet parasitten Bonamia ostreae i østers som var samlet inn i forbindelse med helseovervåking. Cellene ble imidlertid ikke tolket som B. ostreae og PCR-analyser gjort av østers fra denne lokaliteten var negative. Skjellene kom fra en vill bestand nær Arendal i Agder. I 2008 sendte Veterinærinstituttet prøver til EUs referanselaboratorium (EURL). Ved EURL observerte man ved mikroskopering en Bonamia-celle i en østers. Ved real-time PCR-analyse fikk de positivt signal fra to østers (en av triplikate prøver). Sekvensering ga match med Bonamia ostreae. Etter denne diagnosen har både Veterinærinstituttet og Havforskningsinstituttet overvåket bestanden. Resultatene fra dette arbeidet er summert i rapporten. De mikroskopiske cellene er jevnlig observert i prøver fra denne lokaliteten, men alltid med lav prevalens og intensitet. Det er ikke observert inflammasjon eller patologi forbundet med funnene. Østersen er i god kondisjon, bestanden reproduserer som normalt og det er ikke observert tegn til forhøyet dødelighet. Siden 2009 er det analysert mer enn 3 000 østers fra denne lokaliteten. Bonamia sp. er ikke påvist, verken ved histologi eller PCR.
Summarizing the screening for Bonamia ostreae in Norwegian populations of flat oysters, Ostrea edulis.
Rapportserie:
Rapport fra havforskningen 2020-24
ISSN: 1893-4536
Publisert: 31.07.2020
Oppdatert: 08.07.2024
Prosjektnr: 15216
Oppdragsgiver(e): Mattilsynet
Forskningsgruppe(r):
Pathogen Transmission and Disease
Tema:
Flat oyster
Program:
Environmental Impacts of Aquaculture
Approved by:
Research Director(s):
Karin Kroon Boxaspen
Program leader(s):
Terje Svåsand
English summary
Sammendrag
Surveillance and studies performed by The Norwegian Institute of Marine Research and the Norwegian Veterinary Institute has never revealed notifiable diseases in Norwegian populations of European flat oysters, Ostrea edulis. In 2006, microcells resembling the oyster parasite Bonamia sp. were observed during histopathological examination of tissue specimens of flat oysters, Ostrea edulis from the Arendal area, southern Norway. The cells were however not interprated as B. ostreae and PCR-analysis of samples from this oyster population were negative. In 2008, the EU reference laboratory (EURL) received samples from the Norwegian Veterinary Institute and reported one Bonamia sp. in a haemocyte from one oyster, based on microscopy. By real-time PCR, positive results were obtained from two oysters in one triplicate sample. Sequencing of the PCR products gave 100% identity with B. ostreae. After this diagnose, both the Norwegian Veterinary Institute and The Institute of Marine Research have monitored the population. The results are briefly reported here. The observed microcells were found in most samples since the sampling at the site was initiated, always at a low prevalence and intensity. No inflammation, pathology or reductions of the oyster's condition have been associated with the observation. The population appears healthy, with a normal reproductive cycle pattern. Several year classes have been present throughout the study period. Since 2009, more than 3 000 oysters have been examined. Bonamia sp. has never been detected, and all PCR assays have been negative.
1 - Background
The European flat oyster is indigenous to Norwegian coastal waters. Its distribution and abundance is limited by climatic conditions, and has varied considerably in modern time. There are relatively good records dating back to the 1800’s. Oyster farming in Norway was established in 1879 and had its “golden age” until the 1930’s. In this period, oysters were moved and exchanged between some of the Norwegian, Danish and Dutch farm and cultivation sites. There have been no known introductions after this period. The present farming of flat oysters is limited to a few farms, and there is a small-scale commercial harvest from wild stocks. Some wild Norwegian populations are presumably naïve. The Institute of Marine Research monitor the stocks and carries out a health surveillance programme.
2 - Health surveillance of Norwegian oysters
The first health surveillance of farmed bivalve mollusks in Norway was carried out between 1989 and 1992 as a part of a research project ( Mortensen 1993 ) and included flat oysters from two of the main oyster farming sites (Espevik and Vågstranda). Bonamia sp. was not detected in this study.
In 1995, we did a thorough mapping of the remaining oyster farms and farm sites with viable populations. We also mapped some of the oyster beds in southern and western Norway. The results were used as a background for the first official surveillance program for the notifiable oyster parasites Bonamia sp. and Marteilia refringens in Norwegian flat oysters, initiated by the Norwegian Food Safety Authority in 1995. The surveillance included several oyster farms, as well as wild beds. There has been a close contact between the oyster farmers and personnel running the surveillance program in order to ensure that any abnormal mortality was reported. No abnormal mortality has been reported, and neither Bonamia sp. nor Marteilia refringens have been detectedin the flat oysters. In 2004, Norway was officially declared a Bonamia and Marteilia free zone, based on historical data.
The sampling strategy, including wild beds and bivalve farms in operation, was revised in January 2015, and used as a background for the targeted surveillance from 2016, also including blue mussels. Sampling periods were defined according to the periods when the highest prevalence of Bonamia ostreae and Marteilia sp. (spores) have been detected in the northernmost areas where they are present ( Engelsma et al. 2010; A. Alfjorden pers.comm ). Bonamia spp. has never been observed. Possible microcells were observed in oysters from two sites (see below). Marteilia refringens Type M / M. pararefringens was detected in mussels, Mytilus sp. for the first time in 2016.
3 - The Bonamia ostreae diagnose in 2008 – 2009
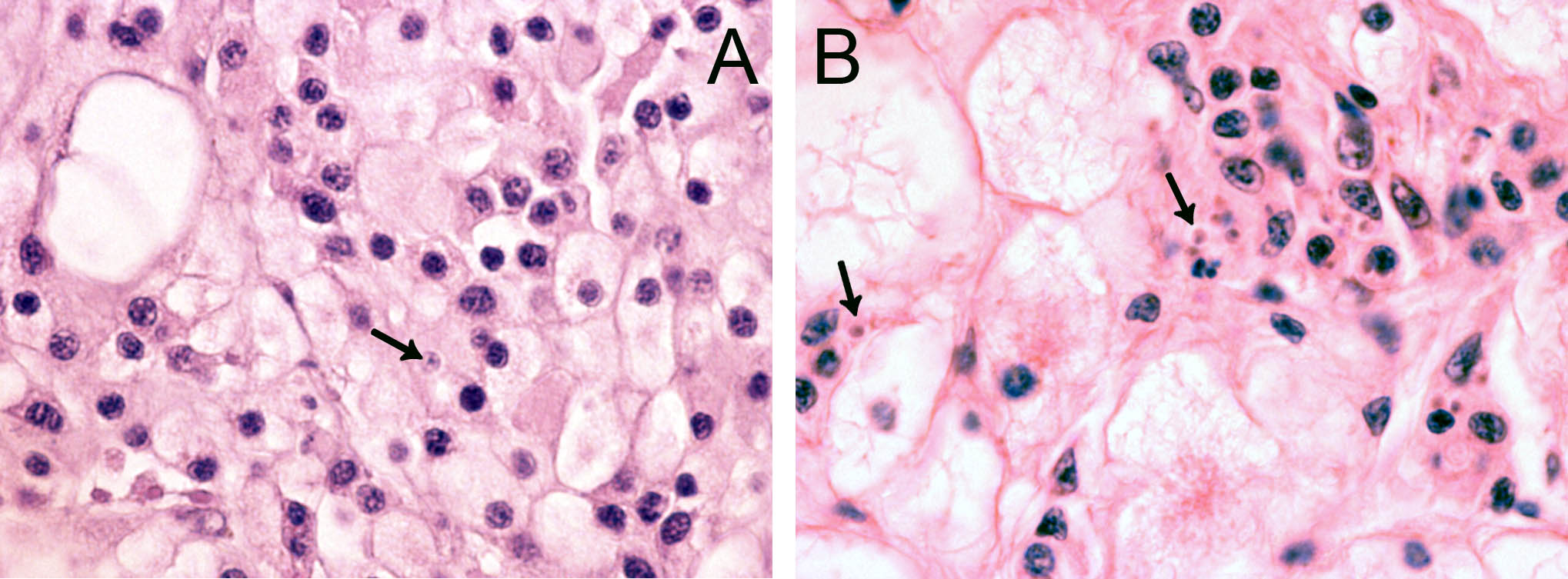
In 2006, cells resembling Bonamia sp. were observed during histopathological examination of tissue specimens of flat oysters, Ostrea edulis from the Arendal area, southern Norway. The cells were slightly larger than B. ostreae compared to reference samples and had a more centric nucleus ( Figure 1 ). These cells were not interpreted as Bonamia during diagnostic surveys, but as they were interpreted as potential “microcells”, samples were sent from the National Veterinary Institute to the European Community Reference Laboratory for Molluscan Diseases in France, for external examination. Bonamia sp. was not confirmed. Similar observations were made in 2008, and nine new samples were sent to France. This time the reference laboratory reported one Bonamia sp. in a haemocyte from one oyster examined by microscopy. In situ hybridization tests ( Cochennec et al . 2000 ) were negative. By real-time PCR, positive results were obtained from two oysters in one triplicate sample. Sequencing of the PCR products gave 100% identity with B. ostreae .
4 - A targeted study in 2009
After the positive diagnosis, a study was initiated by the Food Safety Authority (Mattilsynet) in June 2009, to follow up the case. A new investigation was performed on the oyster population by the Institute of Marine Research (IMR), as well as at three neighboring populations, in order to try to confirm the diagnosis, determine the potential spreading of the parasite and compare histology and PCR as diagnostic tools. Oysters from a farm at Bømlo, on the Norwegian west coast, were used as a presumably negative control. The sites were inspected by skin-diving. There were no signs of recent or previous mass oyster mortalities that could be related to bonamiosis at the examined sites. 50 oysters from each of the four sites were collected and subjected to standard histopathological examination and PCR, using the BO/BOAS ( Cochennec et al . 2000 ) and Cf/Cr ( Carnegie et al . 2000 ) primers. Microscopy of the collected samples confirmed the histopathological findings observed throughout the previous years. However, Bonamia -like cells coinciding with an infection pattern as described for this parasite were not observed: The microcells were present at a very low prevalence, and when present, always as a few single cells. No propagation, inflammation or pathological changes were observed. The PCRs gave negative or inconclusive results, using the two different assays. The investigations did thus not confirm the Bonamia diagnose.
5 - Surveillance and studies after 2009
The official surveillance program has been continued after 2009, carried out by the Veterinary Institute until 2014, with an increased number of oysters collected from the same site every six months (however with some samplings including less than the required 150 oysters ( Table 1, data from Veterinary Institute annual reports from the surveillance programme). Bonamia sp. was not confirmed. In parallel to the surveillance program, monitoring of the population, histological examinations and PCR analysis were carried out by the Institute of Marine Research (included in Table 1 ). From 2015, the surveillance and control programme has been carried out by IMR.
| 2009 | Spring/summer | VI | 146 oysters | Histology |
| IMR | 200 oysters | Histology | ||
| PCR (Marty et al. 2006) | ||||
| Autumn | VI | 112 | Histology | |
| 2010 | Spring | VI | 144 | Histology |
| Summer | IMR | 50 | Histology | |
| PCR (Marty et al. 2006 & Corbeil et al. 2006) | ||||
| Autumn | VI | 123 | Histology | |
| 2011 | Spring | VI | 150 | Histology |
| Summer | IMR | 50 | Histology | |
| PCR (Marty et al. 2006 & Corbeil et al. 2006) | ||||
| Autumn | VI | 150 | Histology | |
| 2012 | Spring | VI | 100 | Histology |
| Summer | IMR | 50 | Histology | |
| PCR (Marty et al. 2006 & Corbeil et al. 2006) | ||||
| Autumn | VI | 0 | Histology | |
| 2013 | Spring | VI | 98 | Histology |
| Summer | IMR | 50 | Histology | |
| PCR (Marty et al. 2006 & Corbeil et al. 2006) | ||||
| Autumn | VI | 145 | Histology | |
| 2014 | Sp ring | VI | 150 | Histology |
| Summer | IMR | 19 | Histology | |
| PCR (Marty et al. 2006 & Corbeil et al. 2006) | ||||
| Autumn | VI | 147 | Histology | |
| 2015 | spring | IMR | 150 | Histology |
| PCR (Marty et al. 2006) | ||||
| autumn | 150 | Histology | ||
| winter | 20 | PCR on haemocytes (Marty et al . 2006) | ||
| 2016 | spring | 150 30 | PCR on haemocytes | |
| autumn | 148 20+20 | PCR on haemocytes | ||
| 2017 | spring | 150 30 | ||
| autumn | 30 | |||
| 2018 | Spring | 150 (20) 30 | Not yet examined. Histo & PCR (30) spring | |
| autumn | 30 | |||
| 2019 | spring | 150 | Not yet examined. | |
| Total | 3042 |
6 - Surveillance in 2015 and 2016 included targeted PCR on haemocytes
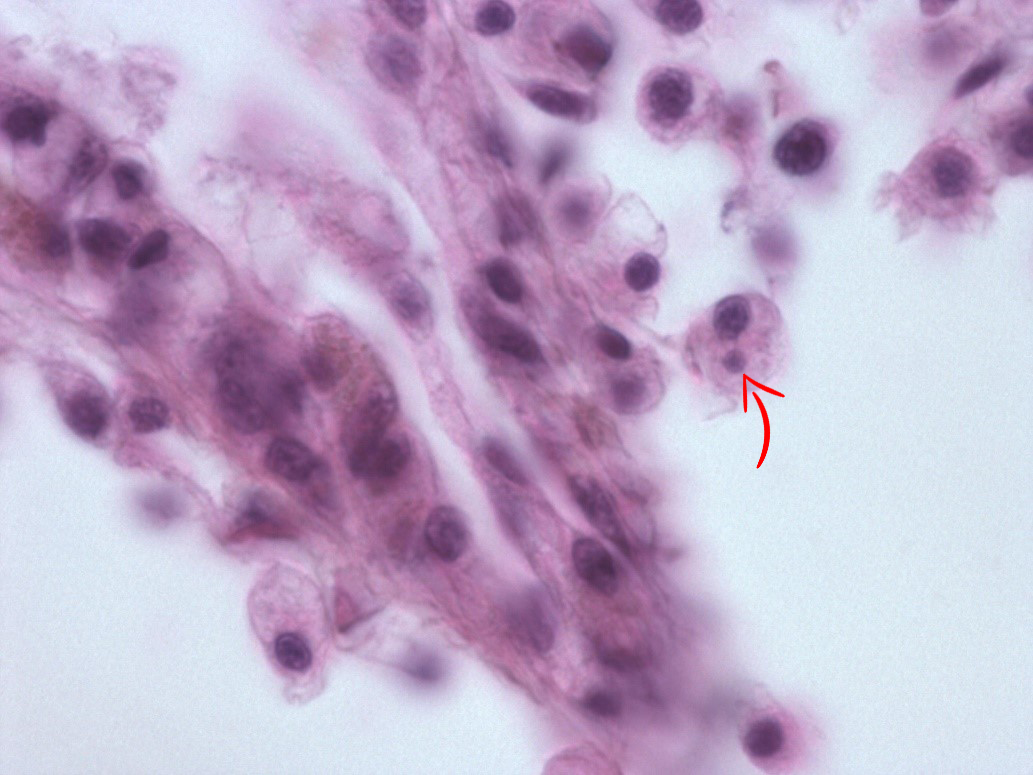
In May 2015, we re-examined the site. There was no sign of abnormal mortality, and several age groups present. 150 flat oysters were collected, processed by standard methods and examined by histology. Microcells were observed as previously, with a prevalence of 10 % and a very low intensity ( Figure 2 ), where cells always appeared individually, and in very low numbers. No inflammation or pathological alterations were observed, and the oysters appeared in good health. Real-time PCR ( Marty et al . 2006 ) was performed on all 150 specimens. All samples were negative, while positive and negative controls gave expected results. The sampling of 150 oysters was repeated in October. There was no sign of abnormal mortality. Microcells were not observed during the histological examination. The oysters appeared in good health. After analyzing the samples from spring 2015, and due to the combination of the microcells observed and the negative PCR results, we collected 20 oysters at 25. November, thus after the autumn sampling ( Table 1 ). In order to obtain a higher number of target cells for the observed microcells, approximately 2 ml haemolymph was withdrawn from the adductor muscle of each oyster. Haemocytes were pelleted, DNA isolated and tested for Bonamia sp. by PCR using the Bo/Boas ( Cochennec et al . 2000 ) and BON-319F/BON-524R ( Hill et al . 2010 ) primers, and the real-time PCR as described above ( Marty et al . 2006 ). All samples were negative, while positive and negative controls gave expected results.
Due to the combination of the microcells observed in 2015 and the negative PCR results, we collected extra haemolymph samples from Langestrand (Agder, see fig. 3) in 2016. We collected 30 samples in May, 20 samples in September and 20 samples in October. In order to obtain a higher number of target cells for the observed microcells, approximately 2 ml haemolymph was withdrawn from the adductor muscle of each oyster. Haemocytes were pelleted, DNA isolated and tested for Bonamia sp. real-time PCR as described above ( Marty et al . 2006 and Corbeil et al 2006 ). All samples were negative.
Additionally, a Microcytos sp. real-time PCR ( Polinski et al . 2015 ) was performed on DNA from the haemocyte samples. Positive controls were kindly provided by Gary Meyer at Virginia Institute of Marine Science. All samples were negative, while positive and negative controls gave expected results.
The Veterinary Institute provided new sections cut from the two original paraffin blocks containing tissues from the two Bonamia positive oysters from 2008. Ten serial sections from each oyster were examined at IMR. Observations were in accordance with all previous samples: cell structures that could represent microcells were observed, but images were not perfectly clear and difficult to interpret. No inflammations or pathological alterations were observed.
7 - Examination of oyster samples from other sites
In addition to the samples from Langestrand, IMR has followed some of the oyster production sites more or less regularly since 1989. Much of the work has focused on the health of flat oysters in some of the farming units producing oyster seed. Bonamia spp. has never been observed. Some of the results from the health surveillance is not published. However, several sites were included in the Surveillance programme for bonamiosis and marteiliosis (National Veterinary Institute until 2014 and IMR since 2015). Bonamia spp. has never been detected.
Samples collected and analyzed by IMR during the Surveillance programme (2015 – present) are listed in Table 2, below.
| Site | 2015 | 2016 | 2017 | 2018 | 2019 | |||||
| spring | autumn | spring | autumn | spring | autumn | spring | autumn | spring | autumn | |
| Hui | 30 | 30 | ||||||||
| Hafrsfjord | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | |
| Sveio | 30 | 30 | 30 | 30 | 30 | 30 | 30 | |||
| Aga | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | ||
| Innerøyen | 30 | |||||||||
8 - Discussion
The shoreline and shallow water area at Langestrand inhabit one of the densest flat oyster populations found in Norway. It is presumed to be naïve, and there are no oyster farms in the area. In order to protect this population, it has been protected from commercial as well as recreational harvest since 2012. IMR has monitored the stock and collected samples for histopathological examination every spring since 2012. No sign of diseases affecting the health of the oysters have been observed. The population appears healthy, with a normal reproductive cycle pattern. Several cohorts have been present throughout the study period.
However, structures resembling microcells have been observed since the sampling at the site at Langestrand was initiated, always at a low prevalence and intensity. No inflammation, pathology or reductions of the oyster's condition have been associated with the observation. The microcells were never interpreted as Bonamia sp. and the diagnose made by EURL was surprising.
Since 2009, more than 3 000 oysters have been examined, and samples from more than 600 of these have been analyzed by PCR, all with negative results. The situation has thus been stable since 2006. A 10 years long sub-clinical Bonamia infection seems unlikely, taking into account that this oyster bed experiences extremely variable conditions through the seasons.
The observed microcells are slightly larger than B. ostreae compared to reference samples and have a more centric nucleus ( Figures 1 & 2 ), resembling Bonamia exitiosa . These should however have been detected by the real-time PCR used. Due to the size and central nucleus, the cells could also be interpreted as Microcytos mackini . The M. mackini PCR however also turned out negative, and the localization of the cells did not correspond to a classical Microcytos detection: The observed cells are always detected in haemocytes, in contrast to Microcytos , which are normally found in connective and muscular tissues.
Samples with the observed microcells were also included in a study of microbiota in flat oysters from Norwegian sites, as a part of the VIVALDI project WP1 (https://cordis.europa.eu/project/id/678589). PCR assays for Bonamia spp, mikrocytids, and paramyxids were all negative. A metagenomic shotgun sequence library was produced from the single available sample, but no candidate parasite sequences were detected. The only positive PCR assay was for haplosporidia, and the products were sequenced. Preliminary phylogenetic analyses showed this to be an early branching haplosporidian. Further work is currently being undertaken to determine whether this sequence derives from the observed microcell.
The nature of the observed cells is thus unresolved. The cells are not believed to be closely related to Bonamia ostreae . If there were sufficient DNA present in any of the samples analyzed by PCR, the known Bonamia species – including B. exitiosa - should have been detected. Other haplosporidian parasites should have been detected by one of the assays applied ( Cochennec et al . 2000 ) on the gill samples in 2009 and the haemocyte samples. In this context, the original diagnose remains a mystery. The haplosporidian sequence generated as described above is currently being further investigated.
9 - Acknowledgements
Thanks to Ingrid U. Fiksdal and Anne Torsvik for technical assistance, Anders Jelmert for assistance with sampling, Cecilie Walde for providing samples from archived samples from the Veterinary institute and Gary Meyer at Virginia Institute of Marine Science for providing positive Microcytos material. Thanks to David Bass (CEFAS, UK) for his contribution with the metagenomic shotgun sequencing.
10 - References
Carnegie, R. B., Barber, B. J., Culloty, S. C., Figueras, A. J., Distel, D. L. (2000). Development of a PCR assay for detection of the oyster pathogen Bonamia ostreae and support for its inclusion in the Haplosporidia. Diseases of Aquatic Organisms 42:199 206.
Cochennec, N., Le Roux, F., Berthe, F., Gerard, A. (2000). Detection of Bonamia ostreae based on small subunit ribosomal probe. Journal of Invertebrate Pathology 76: 26-32.
Corbeil, S., Arzul, I., Robert. M., Berthe, F.C.J., Besnard-Cochennec, N., Crane, M.S.J. (2006). Molecular characterization of an Australian isolate of Bonamia exitiosa . Diseases of Aquatic Organisms 71:82-85.
Engelsma, M.Y., Kerhoff, S., Roozenburg, I., Haenen, O.L.M., van Gool, A., Sistermans, W., Wijnhoven, S., Hummel, H. (2010). Epidemiology of Bonamia ostreae infecting European flat oysters Ostrea edulis from Lake Grevelingen, The Netherlands. Marine Ecology Progress Series 409: 131 – 142.
Hill, K.M., Carnegie, R.B., Aloui-Bejaoui, N., Gharsalli, R., White, D.M., Stokes, N.A., Burreson, E.M. (2010). Observation of a Bonamia sp. Infecting the oyster Ostrea stentina in Tunisia, and a consideration of its phylogenetic affinities. Journal of Invertebrate Pathology 103: 179-185.
Marty, G.D., Bower, S.M., Clarke, K.R., Meyer, G., Lowe, G., Osborn, A.L., Chow, E.P., Hannah, H., Byrne, S., Sojonky, K., Robinson, J.H. (2006). Histopathology and a real-time PCR assay for detection of Bonamia ostreae in Ostrea edulis cultured in western Canada. Aquaculture 261: 33-42.
Mortensen, S.H. (1993). A health survey of selected stocks of commercially exploited Norwegian bivalve molluscs. Diseases of Aquatic Organisms 16: 149-156.
Polinski, M., Lowe, G., Meyer, G., Corbeil, S., Colling, A., Caraguel, C., Abbott, C.L. (2015). Molecular detection of Microcytos mackini in Pacific ousters using quantitative PCR. Molecular & Biochemical Parasitology 200: 19-24.